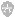酿酒酵母非发酵代谢调控
2007-06-17 14:48:04 来源:网络数据库 评论:0 点击:
Introduction: utilization of carbon sources by Saccharomyces cerevisiae
Among various carbon substrates which can be utilized by the yeast Saccharomyces cerevisiae as a source of cellular biomass and metabolic energy (Barnett 1976), easily fermentable monosaccharides such as glucose or fructose are clearly preferred. In addition to the glycolytic pathway, some specific enzymes are required to metabolize the alternative sugars galactose, sucrose and maltose. Consequently, regulation by the carbon source of GAL, SUC and MAL structural genes has been studied in detail (reviewed by Carlson 1987; Needleman 1991; Bhat and Murthy 2001). Especially, the mechanism of GAL gene expression can now be considered as a paradigm of transcriptional regulation in eukaryotes (Ptashne and Gann 2002). While the presence of galactose as an inducer
of gene expressionmay be a special regulatory event, yeast cells must routinely utilize nonfermentable carbon sources, such as ethanol and glycerol, which are produced when glucose is available in excess. Thus, a hierarchy of regulatory networks evolved, controlling the structural genes which are needed to metabolize various sugars and nonfermentable compounds (glucose repression, carbon catabolite repression). Recent results showed the importance of the respiratory metabolism for extending the lifespan of S. cerevisiae cells (Lin et al. 2002). With cells containing a functional SIR2 gene, growth in 0.5% glucose increased lifespan by about 25%, compared with
cultivation in2%glucose. Thus, a shift from fermentation to a partially respiratory growth causes caloric restriction
which positively affects longevity. This review describes the mechanisms of genetic control required for adaptive utilization of nonfermentable carbon sources by S. cerevisiae. Together with a consideration of carbon flow and the genes/enzymes affected by the carbon source, the current knowledge on signal transduction, regulatory factors, DNA-binding proteins and their recognition of specific sequence motifs which finally lead to transcriptional repression, derepression or induction are presented. Although transcriptional control is in the focus, a distinct mechanism of carbon source regulation (stability of individual mRNAs) is also considered. Related reviews have been published by Ronne (1995), Entian and Schu¨ ller (1997), Gancedo (1998) and Carlson (1999). Nonfermentable substrates must be used for the oxidative metabolism of mitochondria (ATP production) and for the regeneration of sugar phosphates(gluconeogenesis), being substrates of nucleotide biosynthesis, glycosylation, cell wall generation and the biosynthesis of storage carbohydrates. Glycerol entersthe glycolytic pathway at the level of triose phosphate (dihydroxyacetone phosphate; DHAP), which is converted into pyruvate to some degree but also allows production of glucose-6-phosphate. Following its uptake and oxidation to pyruvate, a similar metabolic flow is effective for lactate. Routinely, ethanol produced in the course of alcoholic fermentation is re-introduced into the metabolism via its oxidation and the subsequent biosynthesis of acetyl-CoA. Acetyl-CoA is also generated directly by the peroxisomal b-oxidation of fatty acids. Metabolic pools of acetyl-CoA are required for various pathways, among which the cytoplasmic glyoxylate cycle and the mitochondrial tricarboxylic acid
(TCA) cycle finally lead to sugar phosphates and energy production, respectively (summarized in Fig. 1). Dependent on the carbon substrate, acetyl-CoA and other metabolites are the subject of intracellular transport
which may be controlled by the differential biosynthesis of the respective permeases. Thus, several pathways with partially opposite metabolic fluxes require a complex pattern of gene expression, allowing optimal adaptation to the most convenient substrate available in a certain situation. Consequently, distinct upstream activation sites (UAS) are involved in the transcriptional regulation of structural genes required for respiration, retrograde control of nuclear genes in the case of mitochondrial dysfunction, gluconeogenesis and oleate induction of peroxisomal functions (summarized in Table 1). Activator proteins which bind to these sequences strongly differ with respect to molecular architecture (cf. Fig. 2).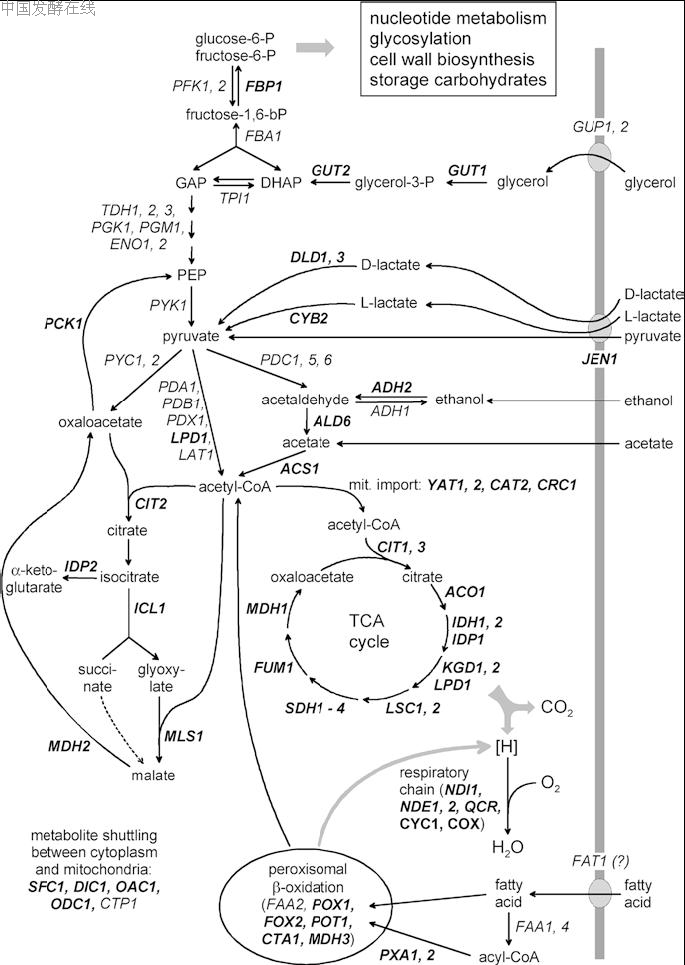
Fig. 1 Genetics of oxidative and gluconeogenic metabolism in Saccharomyces cerevisiae. Structural genes known to be transcriptionally regulated by the carbon source are depicted in bold. The figure does not consider metabolic compartmentation.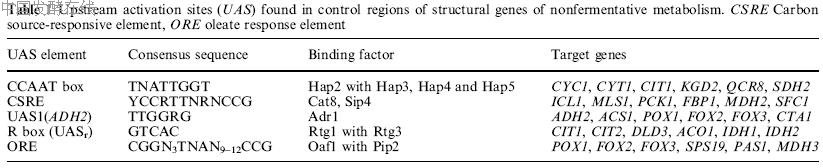
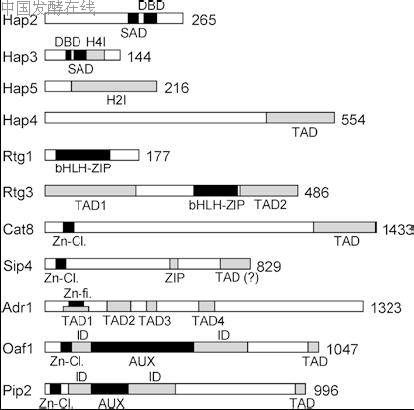
Fig. 2 DNA-binding factors and transcriptional activator proteins of structural genes involved in nonfermentative metabolism of S. cerevisiae. Due to substantial length variations (lengths are given in amino acid residues, at the right), different regulators are not shown to the same scale. As a result of the varying resolution of molecular an
相关热词搜索:Gene regulation Transcriptional acti
上一篇:毕赤酵母表达系统
下一篇:微生物发酵处理对豆粕抗营养因子的影响
评论排行
- ·中国发酵企业数据库(4)
- ·(4)
- ·CoQ10高产菌株选育的研究进展(2)
- ·抗生素发酵工艺所用冷却塔的性能分析及处理(1)
- ·微生物菌种选育技术.rar(1)
- ·发酵生产染菌及其防治(1)
- ·赤藓糖醇发酵工艺研究(1)
- ·重组AiiA 蛋白可溶性表达及发酵条件优化(1)
- ·生物反应器设计软件_发酵罐绿色版(1)
- ·酵母粉、酵母浸粉的区别(1)
- ·雷帕霉素研究进展(1)
- ·透明质酸用途和行业概况(1)
- ·黄酒制作工艺(1)
- ·水解(酸化)工艺与厌氧发酵的区别(1)
- ·糖蜜酒精废液处理过程中产生的微生物蛋...(1)
- ·紫杉醇高产菌发酵产物的分离、纯化和鉴定(1)