酿酒酵母非发酵代谢调控
2007-06-17 14:48:04 来源:网络数据库 评论:0 点击:
uired for DNA binding, transcriptional activation is mediated exclusively by Rtg3 (Rothermel et al. 1997). This conclusion was derived from the finding that activation of GAL1-lacZ by a Gal4-Rtg1 fusion required functional RTG2 and RTG3 genes, while Gal4-Rtg3 could activate even in an rtg1 and rtg2 background. Rtg3 contains two separate activation domains which map to the N-terminus and C-terminus, respectively, of the protein. A central serine/threonine-rich domain of Rtg3 turned out as inhibitory to transcriptional activation.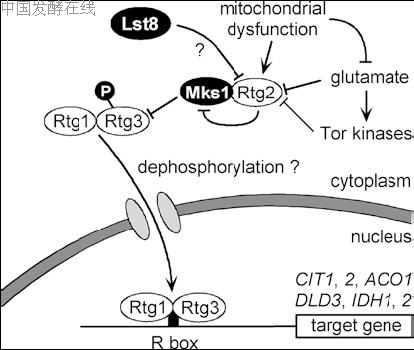
Fig. 5 Regulatory pathway of retrograde control. Signal-dependent nuclear translocation of the Rtg1+Rtg3 heterodimer is required for activation of R box-dependent target genes. Mks1 acts as an inhibitor of Rtg3, possibly by preventing nuclear translocation of the activator complex. Rtg2 counteracts inhibition by Mks1 by partial dephosphorylation of Rtg3. Rtg2 acts as a sensor of respiratory deficiency and availability of glutamate (both signals may coincide). Tor kinases may also act via Rtg2. The function of the negative factor Lst8 is poorly understood. P Phosphate residue, R box regulatory promoter element of retrograde control (upstream activation site UASr)
While expression of CIT2 and DLD3 shows retrograde control and depends on Rtg regulators under all conditions tested, TCA cycle genes CIT1, ACO1, IDH1 and IDH2 require functional RTG genes in cells whose mitochondrial function has been eliminated (Liu and Butow 1999). These additional genes are not affected by retrograde control but are activated by Hap2–Hap
In contrast to R box-binding factors Rtg1 and Rtg3, the function of Rtg2 is less clear. In both q+ and q0 cells, Rtg2 is a cytoplasmic protein with a N-terminal ATPbinding domain and some similarity to bacterial polyphosphatases (according to Liu and Butow 1999). Rtg2 plays a pivotal role as a sensor of the functional state of mitochondria (Komeili et al. 2000; Sekito et al.2000) and also acts in the upstream regulatory pathway of nitrogen catabolism (Pierce et al. 2001). Importantly, activation of RTG-dependent genes correlates with translocation of Rtg1+Rtg3 from the cytoplasm to the nucleus, which requires Rtg2 (Sekito et al. 2000). In rtg2 mutants, Rtg3 is localized constitutively in the cytoplasm, while permanent nuclear localization is found in rtg1 and rtg1 rtg2 mutants. Thus, Rtg1 is both a negative factor of retrograde regulation by retaining Rtg
Rtg2 as a positive factor of retrograde control senses several regulatory inputs (outlined in Fig. 5) and counteracts the Mks1 repressor. Although initially considered as a negative regulator of the Ras-cAMP signaling pathway (Matsuura and Anraku 1993), recent work showed the importance of Mks1 for preventing the nuclear import of Rtg1+Rtg3 (Dilova et al. 2002; Sekito et al. 2002). Mks1 is identical to Lys80, described as a factor required for specific repression of the lysine biosynthetic pathway (Feller et al. 1997). In mks1 and mks1 rtg2 mutant strains, expression of
相关热词搜索:Gene regulation Transcriptional acti
上一篇:毕赤酵母表达系统
下一篇:微生物发酵处理对豆粕抗营养因子的影响
评论排行
- ·中国发酵企业数据库(4)
- ·(4)
- ·CoQ10高产菌株选育的研究进展(2)
- ·抗生素发酵工艺所用冷却塔的性能分析及处理(1)
- ·微生物菌种选育技术.rar(1)
- ·发酵生产染菌及其防治(1)
- ·赤藓糖醇发酵工艺研究(1)
- ·重组AiiA 蛋白可溶性表达及发酵条件优化(1)
- ·生物反应器设计软件_发酵罐绿色版(1)
- ·酵母粉、酵母浸粉的区别(1)
- ·雷帕霉素研究进展(1)
- ·透明质酸用途和行业概况(1)
- ·黄酒制作工艺(1)
- ·水解(酸化)工艺与厌氧发酵的区别(1)
- ·糖蜜酒精废液处理过程中产生的微生物蛋...(1)
- ·紫杉醇高产菌发酵产物的分离、纯化和鉴定(1)