酿酒酵母非发酵代谢调控
2007-06-17 14:48:04 来源:网络数据库 评论:0 点击:
alyses, domains depicted do not necessarily represent functional minimal domains. For zinc-cluster proteins, Cat8, Sip4,Oaf1 and Pip2, middle homology regions (according to Schjerling and Holmberg 1996) are not shown.
AUX Auxiliary domain, bHLH basic helix-loop-helix, DBD DNA-binding domain, H2I (H4I) Hap2 (Hap4) interaction domain, ID inhibitory domain, SAD subunit association domain, TAD transcriptional activation domain, ZIP leucine zipper, Zn-cl. zinc-cluster, Zn-fi. zinc-finger. Data sources are: Hap2–Hap5 (Xing et al. 1993, 1994; McNabb et al. 1995), Rtg1, Rtg3 (Rothermel et al. 1997), Cat8 (Rahner et al. 1996), Sip4 (Lesage et al. 1996), Adr1 (Cook et al. 1994; Chianget al. 1996), Oaf1 and Pip2 (Jia et al. 1997; Baumgartner et al. 1999).
General positive regulators of carbon source utilization Besides several regulatory genes which affect specific pathways of carbon source utilization (discussed below), some controlling factors have a pleiotropic function and
thus influence a large number of target genes. An outstanding regulatory importance has been shown for the snf1 mutation (also known as cat1 or ccr1), which was isolated because of pleiotropic growth defects on media containing sucrose, raffinose, maltose, galactose, glycerol, lactate, ethanol or acetate as the sole carbon source
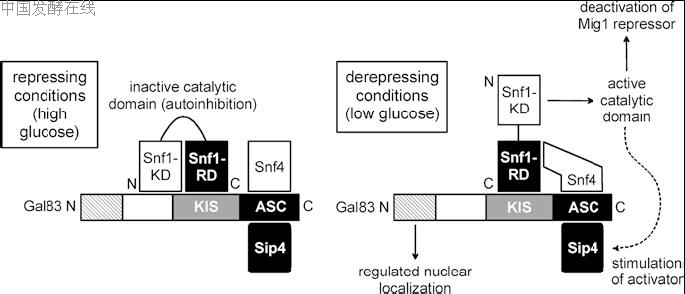
(Ciriacy 1977; Zimmermann et al. 1977; Carlson et al.1981). An identical phenotype was observed for snf4(cat3) mutants (Entian and Zimmermann 1982; Neigeborn and Carlson 1984; Schu¨ ller and Entian 1987).Thus, SNF1 and SNF4 are essential for the metabolic switch occurring after glucose exhaustion and prior to the derepression of glucose-repressible structural genes which are needed for adaptation to an alternative substrate.SNF1 encodes a Ser/Thr-specific protein kinase (Celenza and Carlson 1986, 1989) with high similarity to mammalian AMP-activated protein kinases (AMPK;Carling et al. 1994; Mitchelhill et al. 1994; Woods et al.1994; reviewed by Hardie et al. 1998) and various plant protein kinases, such as RKIN1 which is able to functionally complement a snf1 mutation when expressed in S. cerevisiae (Alderson et al. 1991). AMPK and Snf1 recognize similar target sequences, at least in vitro with the use of model peptides as artificial substrates (FXRXXSXXXF identified as a consensus sequence, where F represents hydrophobic residues L, I, V, F or Mand the phosphorylated serine is shown in italics; Dale et al. 1995). However, Snf1 should be also able to modify unrelated sequences. Recent results identified Snf1 as a histone H3 kinase phosphorylating serine-10 (Lo et al. 2001) within a sequence context which does not fit the consensus motif shown above. Functional dissection of Snf1 could separate its N-terminal catalytic kinase domain (KD) from an inhibitory C-terminal regulatory domain (RD). Importantly, the KD and RD
of Snf1 interact under repressing conditions (high glucose)but not under derepressing conditions, arguing for a carbon source-regulated autoinhibition of the protein kinase (Jiang and Carlson 1996). The SNF4 gene product
is also related to several proteins of mammalian and plant origin (Stapleton et al. 1994) and contains four weakly conserved repeats of the cystathionine-b-synthase domain (Bateman 1997). However, the importance of these sequence repeats is unknown. Snf4 was localized within the nucleus (Schu¨ ller and Entian 1988), is required for Snf1 kinase activity and, thus, must be considered as a positively acting regulatory subunit of Snf1. Indeed, Snf4 interacts with Snf1 (Celenza and Carlson 1989; Celenza et al. 1989; Fields and Song 1989) and represents the c-subunit of the Snf1 protein kinase complex. Interestingly, removal of the negative RD of Snf1 could partially bypass the need for a functional Snf4 (Celenza and Carlson 1989). Since Snf4 binds to the RD of Snf1 under derepressing conditions (contrary to the KD–RD interaction), the c-subunit may be required for the release of the kinase from autoinhibition (Jiang and Carlson 1996; depicted in Fig. 3).
Within the Snf1 complex, additional alternate b-subunits encoded by the related SIP1, SIP2 and GAL83 genes could be identified (Yang et al. 1992, 1994; Erickson and Johnston 1993). Overexpression of SIP1 could suppress certain growth defects of a snf1 mutant, arguing for the functional importance of b-subunits. Although divergent at their N-termini, Sip1, Sip2 and Gal83 share the central kinase-interacting sequence (KIS) domain and the C-terminal associatio
相关热词搜索:Gene regulation Transcriptional acti
上一篇:毕赤酵母表达系统
下一篇:微生物发酵处理对豆粕抗营养因子的影响
评论排行
- ·中国发酵企业数据库(4)
- ·(4)
- ·CoQ10高产菌株选育的研究进展(2)
- ·抗生素发酵工艺所用冷却塔的性能分析及处理(1)
- ·微生物菌种选育技术.rar(1)
- ·发酵生产染菌及其防治(1)
- ·赤藓糖醇发酵工艺研究(1)
- ·重组AiiA 蛋白可溶性表达及发酵条件优化(1)
- ·生物反应器设计软件_发酵罐绿色版(1)
- ·酵母粉、酵母浸粉的区别(1)
- ·雷帕霉素研究进展(1)
- ·透明质酸用途和行业概况(1)
- ·黄酒制作工艺(1)
- ·水解(酸化)工艺与厌氧发酵的区别(1)
- ·糖蜜酒精废液处理过程中产生的微生物蛋...(1)
- ·紫杉醇高产菌发酵产物的分离、纯化和鉴定(1)