Although sugars are clearly the preferred carbon sources of the yeast Saccharomyces cerevisiae, nonfermentable substrates such as ethanol, glycerol, lactate, acetate or oleate can also be used for the generation
of energy
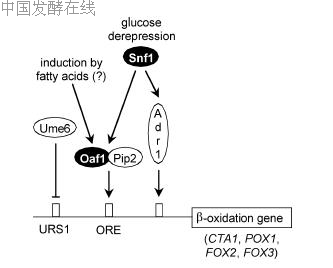
ies of the peroxisomal enzymes involved in b-oxidation increase upon growth with oleate [intensively investigated for the b-keto thiolase gene FOX3 (POT1) and the catalase A gene CTA1]. Transfer of yeast cells from glucose-containing medium to glycerol or ethanol leads to a substantial (2- to 10-fold) derepression of the corresponding genes. In the presence of oleate, gene expression is further induced (2- to 20-fold; Einerhand et al. 1991; Filipits et al. 1993; Karpichev and Small 1998). Both derepression and induction of these genes require a functional Snf1 protein kinase (Simon et al. 1992; Filipits et al. 1993). In contrast, deletion of MIG1 did not alter the regulation of FOX3 (Einerhand et al. 1995). However, FOX3 and some functionally related genes are repressed by Ume6, which binds to URS1 sequence motifs. Interestingly, peroxisomal functions are also affected by retrograde regulation and RTG genes. Loss of mitochondrial DNA (q0) not only leads to an increased transcription of the CIT2 gene encoding the peroxisomal citrate synthase but also stimulates the expression of POX1, FOX2, CTA1,SPS19 and PEX11 (Chelstowska and Butow 1995;Epstein et al. 2001). Consequently, functional RTG genes are necessary for the efficient utilization of oleate. While these data indicate that functional RTG genes may be required for efficient oleate induction, other results argue for a promoter strength which is almost unaffected by a rtg1 mutation (Kos et al. 1995).
Derepression of genes encoding peroxisomal proteins is mediated by Adr1-binding sites (shown for genes POX1, FOX2, FOX3, CTA1, SPS19, PEX1, PEX11;Filipits et al. 1993; Cheng et al. 1994; Simon et al. 1995;Gurvitz et al. 2000, 2001). The transcript level of CTA1,FOX2, FOX3 and PEX1 was clearly reduced in an adr1 mutant, while overexpression of ADR1 could substantially relieve glucose repression of CTA1 transcription (Simon et al. 1991, 1995).
Gene induction by oleate is mediated by a distinct promoter motif, designated oleate response element (ORE; Einerhand et al. 1993; Filipits et al. 1993). From the comparison of sequence variants upstream of boxidation genes, the ORE consensus sequence CGGN3TNAN9–12CCG has been derived (Rottensteiner et al. 1996; Karpichev and Small 1998). In vitro binding studies revealed the existence of a factor specific for interaction with the ORE (Einerhand et al. 1993). Protein binding to the ORE is strongly regulated by the carbon source. While weak interaction was observed with protein extracts prepared from glucose-grown cells, strongly increased signals were obtained with extracts from derepressed or induced cells. Two regulators (Oaf1 and Pip2) which are specific for ORE-dependent gene expression could be identified by purification of the ORE-binding factor and by screening for mutants with a defective oleate induction, respectively (Luo et al. 1996; Rottensteiner et al. 1996). Both Oaf1 and Pip2 contain N-terminal zinc cluster domains (cf. Fig. 2) and exhibit an identity of 39%. Characterization of mutants showed that Oaf1 and Pip2 together are required for oleate induction and for ORE binding. In order to restore ORE-dependent gene activation in an oaf1 pip2 double mutant, both wild-type genes were necessary. Thus, Oaf1 and Pip2 are not alternative activators (such as Cat8 and Sip4 for CSRE-dependent gene expression) but instead bind to the ORE as a heterodimer (shown in Fig. 7; Karpichev et al. 1997; Rottensteiner et al. 1997). Interestingly, OAF1 and PIP2 are differently regulated. PIP2 is positively autoregulated by an ORE in its control region, which mediates a 10-fold induction by oleate in a wild-type strain but not in oaf1 or pip2 mutants (Rottensteiner et al. 1997). In contrast, expression of OAF1 is almost constitutive. No evidence for the regulation of OAF1 or PIP2 by the Mig1 repressor exists. Fusion of full-length Oaf1 or Pip2 to the DNA-binding domain of LexA showed that each protein efficiently mediates transcriptional activation of a LexA-dependent reporter gene, even in the absence of the other (Baumgartner et al. 1999). Activation mediated by both fusion proteins was repressed by glucose and could be induced by oleate. Pip2-dependent activation was fully effective under derepressing conditions, while activation by Oaf1 required inducing conditions. At their C-termini, both Oaf1 and Pip2 contain strong activation core domains which are no longer regulated by the carbon source. Thus, internal auxiliary and inhibitory domains (of. Fig. 2) are required to confer a specific regulatory pattern on both transcription factors. Since its activation specifically occurs under inducing (but not under derepressing) conditions, Oaf1 must be considered as the primary target of fatty acid-mediated transcriptional control. However, the nature of the signal triggering oleate induction is not known. Since ORE-dependent gene expression was also increased in the presence of lauric acid (C12, instead of oleic acid, which is C18), cells do not differentiate between long- and medium-chain fatty acids (Rottensteiner et al. 2002). The signal for induction does not require a functional b-oxidation or biogenesis of intact peroxisomes and is triggered even in the absence of protein synthesis.