酿酒酵母非发酵代谢调控
2007-06-17 14:48:04 来源:网络数据库 评论:0 点击:
re a functional SNF1 gene. Indeed, Cat8 becomes phosphorylated in the course of the derepression of target genes (Randez-Gil et al. 1997). Since Snf1 recognition sites are apparently absent from the sequence, it remains unclear whether Cat8 is a direct substrate of the Snf1 kinase complex. Importantly, a chimeric CAT8 variant with a carbon source-independent promoter and a constitutive transcriptional activation domain fused to Cat8 allowed an almost glucose-insensitive expression of CSRE-driven genes (Rahner et al. 1999). Such a construct could bypass the need for a functional Snf1 protein kinase for growth with ethanol as the sole carbon source. Regulatory mechanisms shown for Cat8 are also effective for the less important CSRE-binding factor Sip4 (Lesage et al. 1996; cf. Fig. 6). SIP4 is responsible for the residual activation of a CSRE-dependent gene in the absence of CAT8 (about 15% of the maximal level of derepression; Hiesinger et al. 2001). Since derepression is completely abolished in a cat8 sip4 double mutant, no additional activators specific for CSRE motifs should exist; and overexpression of either SIP4 or CAT8 could suppress the mutant phenotype of the double mutant. Importantly, Cat8 is required for maximal expression of SIP4 (Vincent and Carlson 1998) and for effective activation by Sip4 (Hiesinger et al. 2001), arguing for some kind of autoregulatory cross-talk among both activators. Indeed, the SIP4 upstream region itself contains a CSRE variant, explaining the reduced transcription of SIP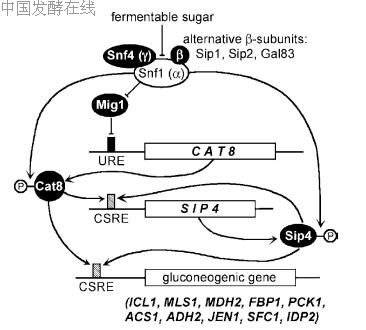
Fig. 6 Derepression of gluconeogenic structural genes by carbon source-responsive element (CSRE)-binding factors, Cat8 and Sip4. A dual function of the Snf1 protein kinase complex is proposed: deactivation of the Mig1 repressor allows biosynthesis of Cat8 and post-translational modification converts Cat8 into a transcriptional activator which subsequently stimulates expression of SIP4. Cat8 and Sip4 both contribute to derepression of gluconeogenic structural genes, with Cat8 being the more important activator. The activator of CAT8 transcription in the absence of Mig1 is unknown. It is also unclear whether Cat8 and Sip4 are directly phosphorylated by Snf1. URE Upstream regulatory element.
Gene regulation by Adr1 Activation of structural genes involved in conversion of C2 substrates into acetyl-CoA requires an additional activator, Adr1, which was initially characterized as a positive regulator of the glucose-repressible alcohol dehydrogenase gene ADH2 (Ciriacy 1975, 1979). The ADH2 control region contains two distinct activating elements, UAS1 and UAS2, both of which individually contribute to carbon source-dependent regulation (Yu et al. 1989).While UAS2 has been recently identified as a CSRE sequence variant and binding site of Cat8 (but not of Sip4; Walther and Schu¨ ller 2001), UAS1 is the target site of Adr1. Consequently, no derepression of ADH2 occurred in an adr1 cat8 double mutant. Adr1 contains two zinc finger motifs of the Cys2-His2 type (Fig. 2;Blumberg et al. 1987; Eisen et al. 1988) and interacts as a monomer with each half-site of the palindromic UAS1 element (Thukral et al. 1991). Functional studies could show that Adr1 monomers recognize the consensus ,sequence TTGGRG (Cheng et al. 1994). However, more efficient activation occurs with two of these motifs in a palindromic orientation, separated by 8–36 nucleotides. Similar to what was shown for ADH2, the combined activities of Adr1 and Cat8 are also required for derepression of the acetyl-CoA
相关热词搜索:Gene regulation Transcriptional acti
上一篇:毕赤酵母表达系统
下一篇:微生物发酵处理对豆粕抗营养因子的影响
评论排行
- ·中国发酵企业数据库(4)
- ·(4)
- ·CoQ10高产菌株选育的研究进展(2)
- ·抗生素发酵工艺所用冷却塔的性能分析及处理(1)
- ·微生物菌种选育技术.rar(1)
- ·发酵生产染菌及其防治(1)
- ·赤藓糖醇发酵工艺研究(1)
- ·重组AiiA 蛋白可溶性表达及发酵条件优化(1)
- ·生物反应器设计软件_发酵罐绿色版(1)
- ·酵母粉、酵母浸粉的区别(1)
- ·雷帕霉素研究进展(1)
- ·透明质酸用途和行业概况(1)
- ·黄酒制作工艺(1)
- ·水解(酸化)工艺与厌氧发酵的区别(1)
- ·糖蜜酒精废液处理过程中产生的微生物蛋...(1)
- ·紫杉醇高产菌发酵产物的分离、纯化和鉴定(1)